What are stem cells and their different types?
Differentiation and senescence
The process through which dividing cells alter their phenotypic or functional type is known as differentiation (Hwang, Zhang, Hwang, & Varghese, 2009). Normal cells can experience cellular senescence, a stable cell cycle arrest, in response to a variety of internal and external stimuli as well as developmental indications (Kumari & Jat, 2021) (Koch et al., 2012).
Role of DNA methylation in cell
DNA methyltransferases (DNMTs) facilitate the transfer of methyl groups to the genome, which methylates DNA and regulates the expression of genes which helps achieve Genome methylation or DNA methylation (Chattopadhyaya & Ghosal, 2022).
This blog refers to a study conducted by (Giuliani et al., 2023):
- To examine the DNA alterations in human-derived (h-)BMSCs committed to osteoblast and adipocyte differentiation as well as replicative senescence.
- Assessing a DNA signature that is common or specific to a certain cell type in the development of replicative senescence.
Genome-wide DNAm analysis (Sugimoto et al., 2020) was performed in BMSCs and HUVECs at early and late passages, as well as in BMSCs induced to differentiate into osteoblasts (OS) and adipocytes (AD), in order to identify DNAm patterns linked to replicative senescence and BMSC differentiation.
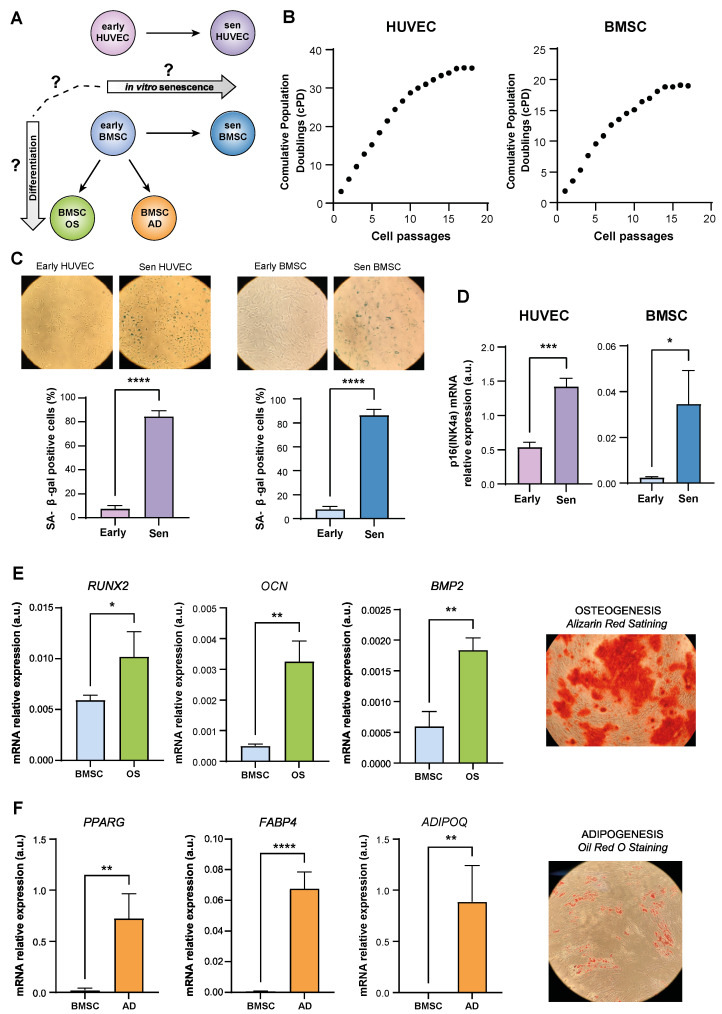
The primary causes of variation in genome-wide DNA across cell type, differentiation, and senescence status were investigated using Principal Component Analysis (PCA).
In BMSCs and HUVECs, a search was conducted for differentially methylated positions (DMPs) linked to senescence. Several one-sample Wilcoxon tests further supported this hypomethylation pattern.
Furthermore, datasets were analyzed to detect DMPs between BMSCs that were undifferentiated and BMSCs that had differentiated into adipocytes and osteoblasts.
Lastly, research was done on the connection between the DNAm modifications that senescent BMSCs and differentiated BMSCs underwent.
In a nutshell, BMSC differentiation and senescence are associated with genome-wide DNA hypomethylation; this hypomethylation, however, involves distinct sets of CpG probes (Fig. S2), allowing distinct pathways that these cells could follow to be distinct. Understanding how the senescence status may affect the differentiation potential of mesenchymal stem cells might be aided by DNA methylation analysis. To translate these results into the study of age-related bone disease, more mechanistic research is required.
References
Chattopadhyaya, S., & Ghosal, S. (2022). DNA methylation: a saga of genome maintenance in hematological perspective. Human Cell, 35(2), 448-461. https://doi.org/10.1007/s13577-022-00674-9
Giuliani, A., Bacalini, M. G., Ramini, D., Mensà, E., Giordani, C., Xumerle, L., Garagnani, P., Olivieri, F., Procopio, A. D., Rippo, M. R., et al. (2023). Genome-wide methylation changes associated with replicative senescence and differentiation in endothelial and bone marrow mesenchymal stromal cells. Cells, 12(2), 285. https://doi.org/10.3390/cells12020285
Hwang, N. S., Zhang, C., Hwang, Y. S., & Varghese, S. (2009). Mesenchymal stem cell differentiation and roles in regenerative medicine. Wiley Interdisciplinary Reviews: Systems Biology and Medicine, 1(1), 97-106. https://doi.org/10.1002/wsbm.26
Koch, C. M., Joussen, S., Schellenberg, A., Lin, Q., Zenke, M., & Wagner, W. (2012). Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell, 11(2), 366-369. https://doi.org/10.1111/j.1474-9726.2011.00784.x
Kumari, R., & Jat, P. (2021). Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Frontiers in Cell and Developmental Biology, 9. https://doi.org/10.3389/fcell.2021.645593
Sugimoto, K., Momose, H., Ito, T., Orita, H., Sato, K., Sakamoto, K., & Brock, M. V. (2020). Genome-wide analysis of DNA methylation in gastrointestinal cancer. Journal of Visualized Experiments, (163), e61355. https://doi.org/10.3791/61355
Total word count: 381
